import xarray as xr
import os,glob
import numpy as np
from pyhdf.SD import SD
path = '/Datadisk/TRMM/3B43/'
file_list = []
for year in range(2009,2019):
folder = os.path.join(path, str(year))
file_name = glob.glob(folder+'/3B43.'+str(year)+'*.HDF')
file_name.sort()
file_list.extend(file_name)
file_list.sort()
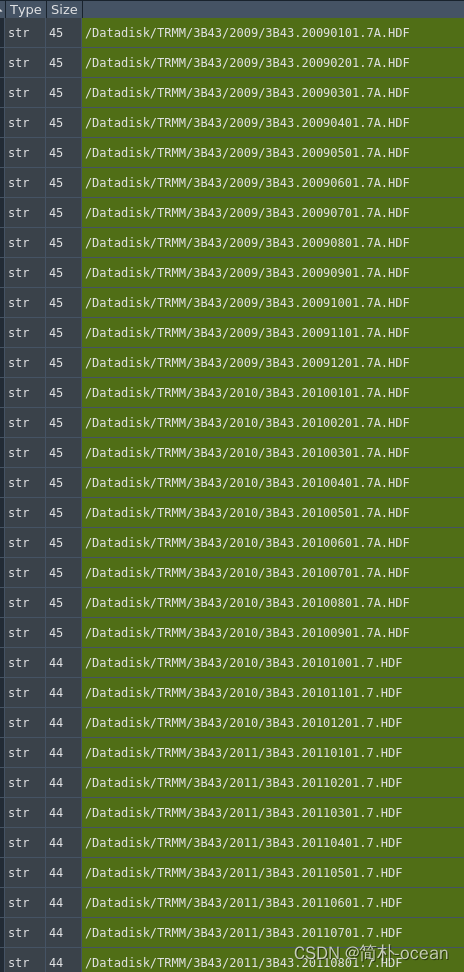
2、封装数据读取函数,并对需要的区域进行切片,我选择的区域为经度:[90.0, 145],纬度:[-10, 55],并将循环读取的10年月平均数组创建为dataarray的格式方面后续掩膜,这里的时间可以通过pandas自己创建时间序列,我这里偷懒直接读取了之前处理过的月平均gpcp的time了
dt = xr.open_dataset("/gpcp_monthly_mask.nc")
precip = dt.sel(time=slice('2009','2018'),lat =slice(-10,55),lon = slice(90,145)).precip
time_num = precip.time.values
def get_data(path,time):
da = SD(path)
pre = da.select('precipitation')[:]
pre = np.expand_dims(pre,axis=0)
lon = np.arange(-180,180.,0.25)
lat = np.arange(-50,50.,0.25)
time = time
da = xr.DataArray(pre,
dims=['time','lon','lat'],
coords=dict(
lon=(['lon'], lon),
lat=(['lat'], lat),
time=(['time'],[time])),
lon_range = [90.0, 145]
lat_range = [-10, 55]
da = da.sel(lon=slice(*lon_range), lat=slice(*lat_range))
x ,y = da.lon,da.lat
return da,x,y
rain = np.zeros((len(file_list),221,240))
for i in range(len(file_list)):
print(i)
da,x,y = get_data(file_list[i], time_num[i])
rain[i] = da
ds = xr.DataArray(rain,
dims=['time','lon','lat'],
coords=dict(
lon=(['lon'], x.data),
lat=(['lat'], y.data),
time=(['time'],time_num)),
3、对数据的陆地部分进行掩膜,并计算气候态平均,转换单位为mm/year
from global_land_mask import globe
def mask_land(ds, label='land', lonname='lon'):
if lonname == 'lon':
lat = ds.lat.data
lon = ds.lon.data
if np.any(lon > 180):
lon = lon - 180
lons, lats = np.meshgrid(lon, lat)
mask = globe.is_ocean(lats, lons)
temp = []
temp = mask[:, 0:(len(lon) // 2)].copy()
mask[:, 0:(len
(lon) // 2)] = mask[:, (len(lon) // 2):]
mask[:, (len(lon) // 2):] = temp
else:
lons, lats = np.meshgrid(lon, lat)
mask = globe.is_ocean(lats, lons)
ds.coords['mask'] = (('lat', 'lon'), mask)
elif lonname == 'longitude':
lat = ds.latitude.data
lon = ds.longitude.data
if np.any(lon > 180):
lon = lon - 180
lons, lats = np.meshgrid(lon, lat)
mask = globe.is_ocean(lats, lons)
temp = []
temp = mask[:, 0:(len(lon) // 2)].copy()
mask[:, 0:(len(lon) // 2)] = mask[:, (len(lon) // 2):]
mask[:, (len(lon) // 2):] = temp
else:
lons, lats = np.meshgrid(lon, lat)
mask = globe.is_ocean(lats, lons)
lons, lats = np.meshgrid(lon, lat)
mask = globe.is_ocean(lats, lons)
ds.coords['mask'] = (('latitude', 'longitude'), mask)
if label == 'land':
ds = ds.where(ds.mask == True)
elif label == 'ocean':
ds = ds.where(ds.mask == False)
return ds
data = mask_land(ds,'land')
precip_mean = np.nanmean(data*24*365,axis=0)
4、绘图,保存图片
import cartopy.feature as cfeature
from cartopy.mpl.ticker import LongitudeFormatter,LatitudeFormatter
import cmaps
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
import matplotlib.ticker as mticker
from cartopy.mpl.gridliner import LONGITUDE_FORMATTER, LATITUDE_FORMATTER
from cnmaps import get_adm_maps, draw_maps
box = [100,140,0,50]
xstep,ystep = 10,10
proj = ccrs.PlateCarree(central_longitude=180)
plt.rcParams['font.family'] = 'Times New Roman',
fig = plt.figure(figsize=(8,7),dpi=200)
fig.tight_layout()
ax = fig.add_axes([0.1,0.2,0.8,0.7],projection = proj)
ax.set_extent(box,crs=ccrs.PlateCarree())
draw_maps(get_adm_maps(level='国'))
ax.set_xticks(np.arange(box[0],box[1]+xstep, xstep),crs=ccrs.PlateCarree())
ax.set_yticks(np.arange(box[2], box[3]+1, ystep),crs=ccrs.PlateCarree())
lon_formatter = LongitudeFormatter(zero_direction_label=False)
lat_formatter = LatitudeFormatter()
ax.xaxis.set_major_formatter(lon_formatter)
ax.yaxis.set_major_formatter(lat_formatter)
ax.set_title('TRMM(mm/year)',fontsize=16,pad=8,loc='left')
ax.tick_params( which='both',direction='in',
width=0.7,
pad=8,
labelsize=14,
bottom=True, left=True, right=True, top=True)
c = ax.contourf(x,y,precip_mean.
T,
levels=np.arange(200,3300,100),
extend='both',
transform=ccrs.PlateCarree(),
cmap=cmaps.NCV_jet)
cb=plt.colorbar(c,
shrink=0.98,
orientation='vertical',
aspect=28,
cb.ax.tick_params(labelsize=10,which='both',direction='in',)
plt.show()
fig.savefig('./TRMM_10year_monthly.png',dpi=500)
Created on Fri May 5 09:45:08 2023
@author: %(jixianpu)s
Email : 211311040008@hhu.edu.cn
introduction : keep learning althongh walk slowly
import xarray as xr
import os,glob
import numpy as np
from pyhdf.SD import SD
path = '/Datadisk/TRMM/3B43/'
file_list = []
for year in range(2009,2019):
folder = os.path.join(path, str(year))
file_name = glob.glob(folder+'/3B43.'+str(year)+'*.HDF')
file_name.sort()
file_list.extend(file_name)
file_list.sort()
dt = xr.open_dataset("/gpcp_monthly_mask.nc")
precip = dt.sel(time=slice('2009','2018'),lat =slice(-10,55),lon = slice(90,145)).precip
time_num = precip.time.values
def get_data(path,time):
da = SD(path)
pre = da.select('precipitation')[:]
pre = np.expand_dims(pre,axis=0)
lon = np.arange(-180,180.,0.25)
lat = np.arange(-50,50.,0.25)
time = time
da = xr.DataArray(pre,
dims=['time','lon','lat'],
coords=dict(
lon=(['lon'], lon),
lat=(['lat'], lat),
time=(['time'],[time])),
lon_range = [90.0, 145]
lat_range = [-10, 55]
da = da.sel(lon=slice(*lon_range), lat=slice(*lat_range))
x ,y = da.lon,da.lat
return da,x,y
rain = np.zeros((len(file_list),221,240))
for i in range(len(file_list)):
print(i)
da,x,y = get_data(file_list[i], time_num[i])
rain[i] = da
ds = xr.DataArray(rain,
dims=['time','lon','lat'],
coords=dict(
lon=(['lon'], x.data),
lat=(['lat'], y.data),
time=(['time'],time_num)),
from global_land_mask import globe
def mask_land(ds, label='land', lonname='lon'):
if lonname == 'lon':
lat = ds.lat.data
lon = ds.lon.data
if np.any(lon > 180):
lon = lon - 180
lons, lats = np.meshgrid(lon, lat)
mask = globe.is_ocean(lats, lons)
temp = []
temp = mask[:, 0:(len(lon) // 2)].copy()
mask[:, 0:(len(lon)
// 2)] = mask[:, (len(lon) // 2):]
mask[:, (len(lon) // 2):] = temp
else:
lons, lats = np.meshgrid(lon, lat)
mask = globe.is_ocean(lats, lons)
ds.coords['mask'] = (('lat', 'lon'), mask)
elif lonname == 'longitude':
lat = ds.latitude.data
lon = ds.longitude.data
if np.any(lon > 180):
lon = lon - 180
lons, lats = np.meshgrid(lon, lat)
mask = globe.is_ocean(lats, lons)
temp = []
temp = mask[:, 0:(len(lon) // 2)].copy()
mask[:, 0:(len(lon) // 2)] = mask[:, (len(lon) // 2):]
mask[:, (len(lon) // 2):] = temp
else:
lons, lats = np.meshgrid(lon, lat)
mask = globe.is_ocean(lats, lons)
lons, lats = np.meshgrid(lon, lat)
mask = globe.is_ocean(lats, lons)
ds.coords['mask'] = (('latitude', 'longitude'), mask)
if label == 'land':
ds = ds.where(ds.mask == True)
elif label == 'ocean':
ds = ds.where(ds.mask == False)
return ds
data = mask_land(ds,'land')
precip_mean = np.nanmean(data*24*365,axis=0)
import cartopy.feature as cfeature
from cartopy.mpl.ticker import LongitudeFormatter,LatitudeFormatter
import cmaps
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
import matplotlib.ticker as mticker
from cartopy.mpl.gridliner import LONGITUDE_FORMATTER, LATITUDE_FORMATTER
from cnmaps import get_adm_maps, draw_maps
box = [100,140,0,50]
xstep,ystep = 10,10
proj = ccrs.PlateCarree(central_longitude=180)
plt.rcParams['font.family'] = 'Times New Roman',
fig = plt.figure(figsize=(8,7),dpi=200)
fig.tight_layout()
ax = fig.add_axes([0.1,0.2,0.8,0.7],projection = proj)
ax.set_extent(box,crs=ccrs.PlateCarree())
draw_maps(get_adm_maps(level='国'))
ax.set_xticks(np.arange(box[0],box[1]+xstep, xstep),crs=ccrs.PlateCarree())
ax.set_yticks(np.arange(box[2], box[3]+1, ystep),crs=ccrs.PlateCarree())
lon_formatter = LongitudeFormatter(zero_direction_label=False)
lat_formatter = LatitudeFormatter()
ax.xaxis.set_major_formatter(lon_formatter)
ax.yaxis.set_major_formatter(lat_formatter)
ax.set_title('TRMM(mm/year)',fontsize=16,pad=8,loc='left')
ax.tick_params( which='both',direction='in',
width=0.7,
pad=8,
labelsize=14,
bottom=True, left=True, right=True, top=True)
c = ax.contourf(x,y,precip_mean.T,
levels=np.arange(200,3300,100),
extend='both',
transform=ccrs.PlateCarree(),
cmap=cmaps.NCV_jet)
cb=plt.colorbar(c,
shrink=0.98,
orientation='vertical',
aspect=28,
cb.ax.tick_params(labelsize=10,which='both',direction='in',)
plt.show()
fig.savefig('./TRMM_10year_monthly.png',dpi=500)
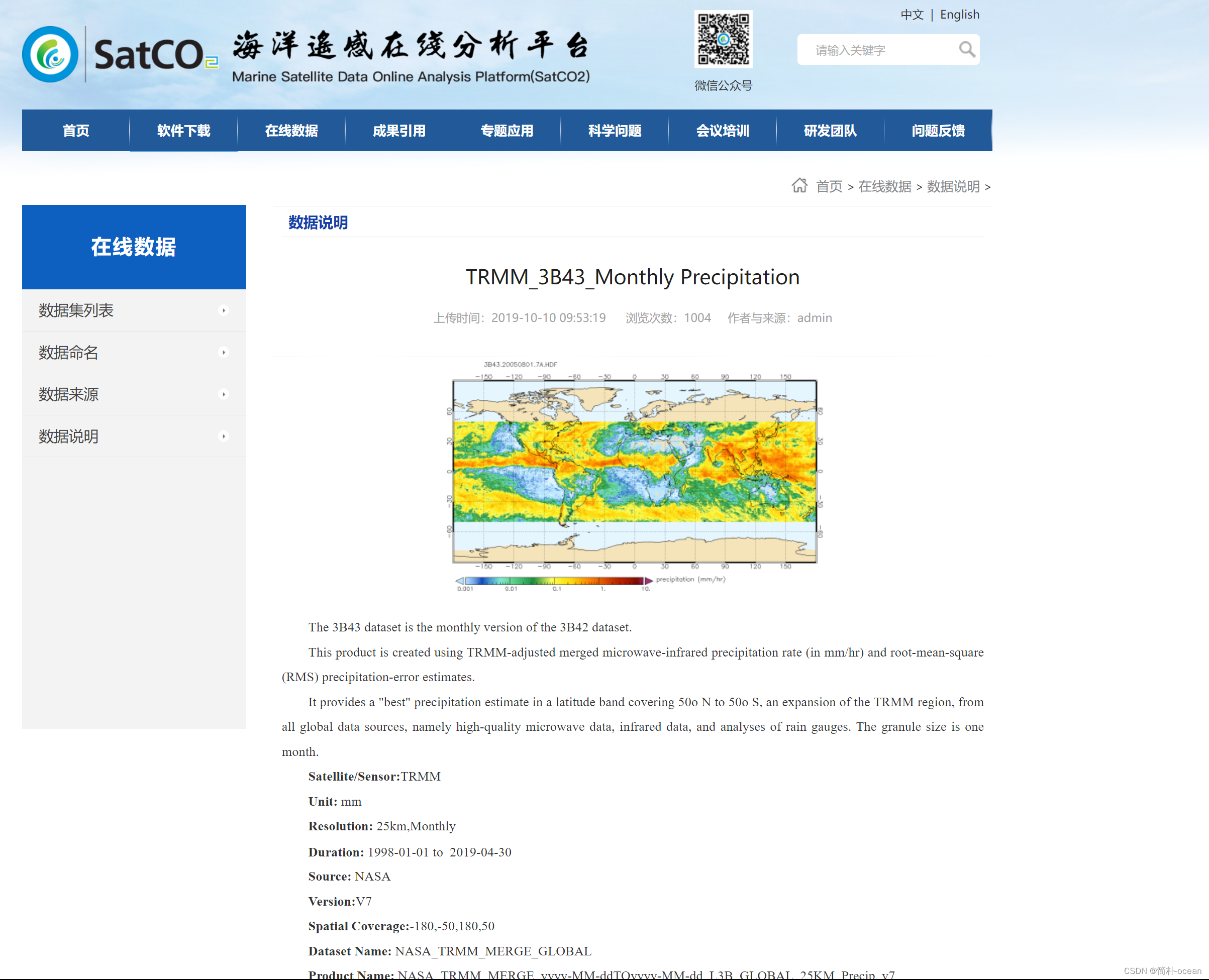
TRMM: Tropical Rainfall Measuring Mission
https://climatedataguide.ucar.edu/climate-data/trmm-tropical-rainfall-measuring-mission
Monthly 0.25° x 0.25° TRMM multi-satellite and Other Sources Rainfall (3B43)
http://apdrc.soest.hawaii.edu/datadoc/trmm_3b43.php

TRMM 3B43: Monthly Precipitation Estimates
https://developers.google.com/earth-engine/datasets/catalog/TRMM_3B43V7
中巴经济走廊TRMM_3B43月降水数据(1998-2017年):http://www.ncdc.ac.cn/portal/metadata/4b9504fa-0e34-47c9-a755-91d3f3253312

TRMM (TMPA/3B43) Rainfall Estimate L3 1 month 0.25 degree x 0.25 degree V7 (TRMM_3B43)(GES 官网介绍):https://disc.gsfc.nasa.gov/datasets/TRMM_3B43_7/summary
国家海洋遥感在线分析平台 https://www.satco2.com/index.php?m=content&c=index&a=show&catid=317&id=217
之前没怎么处理过程HDF的文件,时间仓促,只是简单了记录了一下,没有考虑代码的美观和计算的高效性,欢迎大家评论或者联系我进行交流讨论~~
将TRMM降水数据的处理过程进行梳理,使用Matlab工具获取区域降水异常的变化速率和区域平均值,能够帮助实现与降水的耦合分析,本文所使用的程序以及实例数据均通过了验证。
提示:以下是本篇文章正文内容,下面案例可供参考
一、TRMM降水数据
这里使用的数据为TRMM_3B43数据,是包含降水量的数据集,TRMM全球降雨数据产品由美国国家....
利用多种回归模型对华北八省市的实验区TRMM影像数据进行矫正
回归模型包括:多元线性回归、贝叶斯岭回归、弹性网络回归、支持向量机回归、梯度增强回归
通过对比发现:梯度增强回归模型的拟合效果最好,精度最高
回归模型的自变量包括:TRMM影像数据、DEM数据、经度、纬度
因变量为:实验区的站点插值数据
模型建立之后,将其应用于测试集数据,实现降雨数据的矫正
TRMM数据处理可以使用Matlab进行。首先,我们需要导入TRMM数据文件,这些文件通常是HDF格式的。在Matlab中,可以使用dfread函数来读取HDF文件中的数据。例如,使用以下代码可以读取TRMM数据文件中的降水数据:
precipitation = hdfread('D:\3B43.19980101.7.HDF', '/Grid/precipitation', 'Index', {[1 1],[1 1],[1440 400]});
这行代码将读取名为'3B43.19980101.7.HDF'的文件中的降水数据,并将其存储在名为precipitation的变量中。'/Grid/precipitation'是HDF文件中存储降水数据的路径。'Index'参数指定了要读取的数据的索引范围,这里是从第1行第1列开始,读取一个1x1的数据块,总共读取1440行和400列的数据。
接下来,你可以根据需要对数据进行进一步的处理和分析。例如,你可以计算降水的统计特征,绘制降水分布图等。Matlab提供了丰富的函数和工具箱来处理和分析数据。
引用[1]和引用[2]提供了关于TRMM数据的读取和处理的具体方法和步骤,你可以参考这些引用中的内容来进行TRMM数据的处理。
如何解决 conda install 库时报错:The environment is inconsistent, please check the package plan carefully
14953






