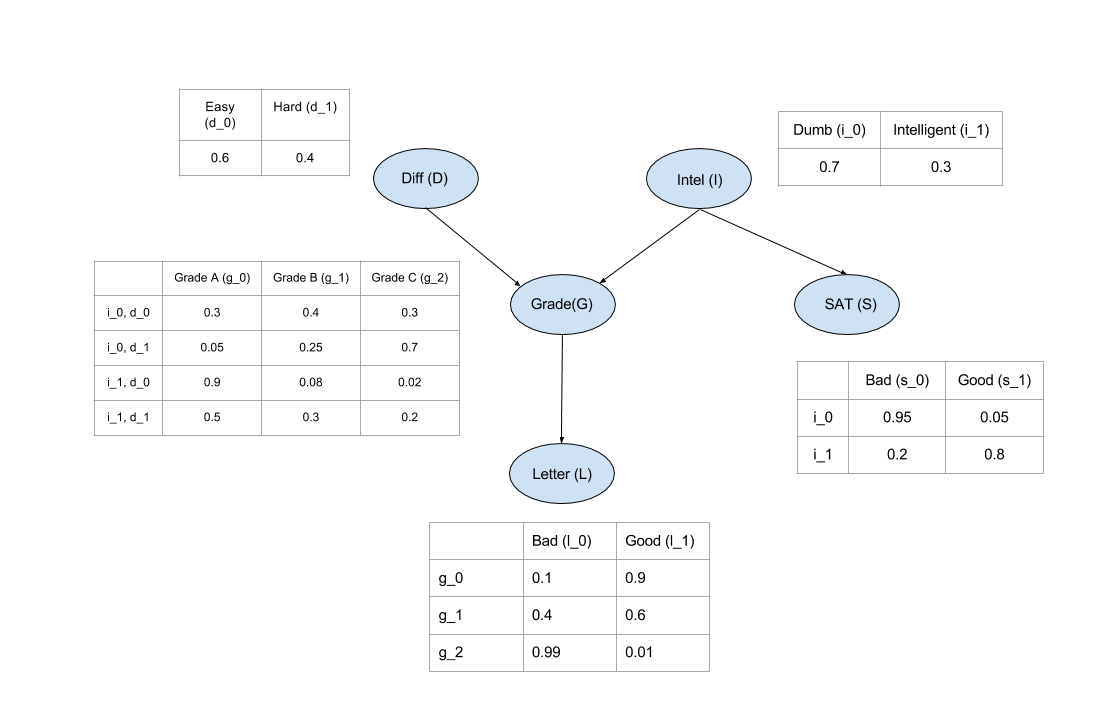
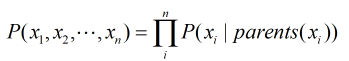Till now we just have been considering that the Bayesian Network can represent the Joint Distribution without any proof. Now let’s see how to compute the Joint Distribution from the Bayesian Network.
Till now we discussed just about representing Bayesian Networks. Now let’s see how we can do inference in a Bayesian Model and use it to predict values over new data points for machine learning tasks. In this section we will consider that we already have our model. We will talk about constructing the models from data in later parts of this tutorial.
In inference we try to answer probability queries(概率查询) over the network given some other variables. So, we might want to know the probable grade of an intelligent student in a difficult class given that he scored good in SAT. So for computing these values from a Joint Distribution we will have to reduce over the given variables that is
P(G∣I=1,D=1,S=1). But carrying on marginalize and reduce operation on the complete Joint Distribution is computationaly expensive since we need to iterate over the whole table for each operation and the table is exponential is size to the number of variables. But in Graphical Models we exploit the independencies to break these operations in smaller parts making it much faster.
One of the very basic methods of inference in Graphical Models is Variable Elimination.
Variable Elimination
由贝叶斯的联合概率:
P(D,I,G,L,S)=P(L∣G)∗P(S∣I)∗P(G∣D,I)∗P(D)∗P(I)
计算G的概率,我们需要边缘化所有其他变量
P(G)=∑D,I,L,SP(L∣G)∗P(S∣I)∗P(G∣D,I)∗P(D)∗P(I)
由局部独立条件化简得:
P(G)=∑
D∑I∑L∑SP(L∣G)∗P(S∣I)∗P(G∣D,I)∗P(D)∗P(I)
Now since not all the conditional distributions depend on all the variables we can push the summations inside:
由于并不是所有的条件分布都依赖于所有变量,所以可以变换一下:
P(G)=∑D∑I∑L∑SP(L∣G)∗P(S∣I)∗P(G∣D,I)∗P(D)∗P(I)
最后将求和推到里面,可以简化大量的计算
P(G)=∑DP(D)∑IP(G∣D,I)∗P(I)∑SP(S∣I)∑LP(L∣G)